Multi-dimensional network scaling for python.
A python module for the multi-dimensional scaling of networks. MDScaling takes topological data (a network of connected nodes) an produces a multi-dimensional embedding in a geometrical space where distance is related to topological similarity.
Cite us:
Ramaciotti Morales, P., Cointet, J.-P., & Laborde, P. (2020, December). Your most telling friends: Propagating latent ideological features on Twitter using neighborhood coherence. In 2020 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM). IEEE.
pip install mdscaling
This is the module's main application. It allows for the embeddings/scaling of directed bipartite graphs, for a range of methods, a range of parameters, and the computation of comparison metrics for the methods x parameters space.
The input format is a CSV file with two columns and no header. Each line is an edge of the bipartite graph. Bipartite graphs have a set of top nodes an a set of bottom nodes. In the input file, the first columns contains the labels of the bottom nodes, and the second column contains the nodes of the top nodes.
For example, the first 5 lines of dataset/twitter_france.csv read
B0,A591
B1,A591
B2,A591
B3,A591
B4,A591
defining 5 edges, connecting bottom nodes (Twitter followers) B0, B1, B2, B3, and B4 with top node A591 (a parliamentarian).
Quick start using datasets from the datasetfolder as
import mdscaling as mds
DB = mds.DiBipartite('local_data/twitter_france.csv') # read the French MPs-followers bipartite graph
DB.CA() # produce an embedding of top and bottom nodes with default values
Embedding coordinates of nodes are then available in the DB.embedding dataframe. The Correspondance Analysis embedding CA allows for specifying some parameters
DB.CA(theta=3,dimensions=3,coordinates='top',all_nodes=False)
coordinates is the part of the bipartite used as original coordinates for the embedding procedure. theta is the threshold of coordinate-nodes-degree to select nodes for which to compute the embedding transformation. This transformation can be compute, nonetheless, to all nodes, by selection all_nodes=True. dimensions allows for specifying the number of dimensions of the embedding.
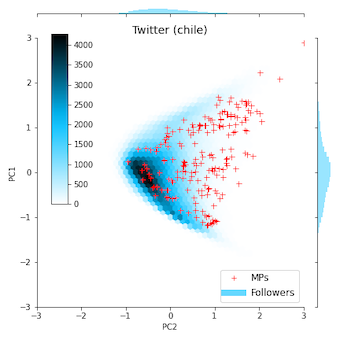
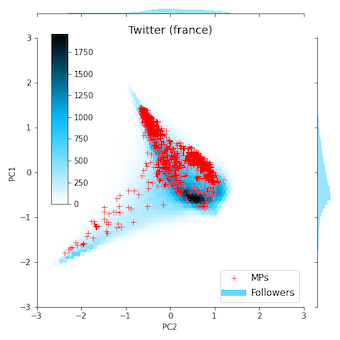
The following two figures show embeddings produced for the Chilean and French Twitter networks available in the datasetsfolder, using the script tests/twitter_example.py.
This is work in progress...